The Spliceosome – new insights into complex molecular machineries
Excision of intron-sequences from RNA-molecules is accomplished by a two step reaction: In a first step an intron-lariat is formed and the first exon is liberated. In a second step the two exons are connected and the intron lariat is released. Galej et al. elucidated the spliceosome structure just after the first catalytic step with the intron lariat/exon 2 and a separated exon1 attached to the complex (see figure above). A structure of an activated spliceosome complex just prior to a final activation step and the subsequent first catalytic step has been provided by Rauhut et al.. Both studies provide detailed insights into the structural changes occuring during preparation and functioning of the eukaryotic splicing machinery.
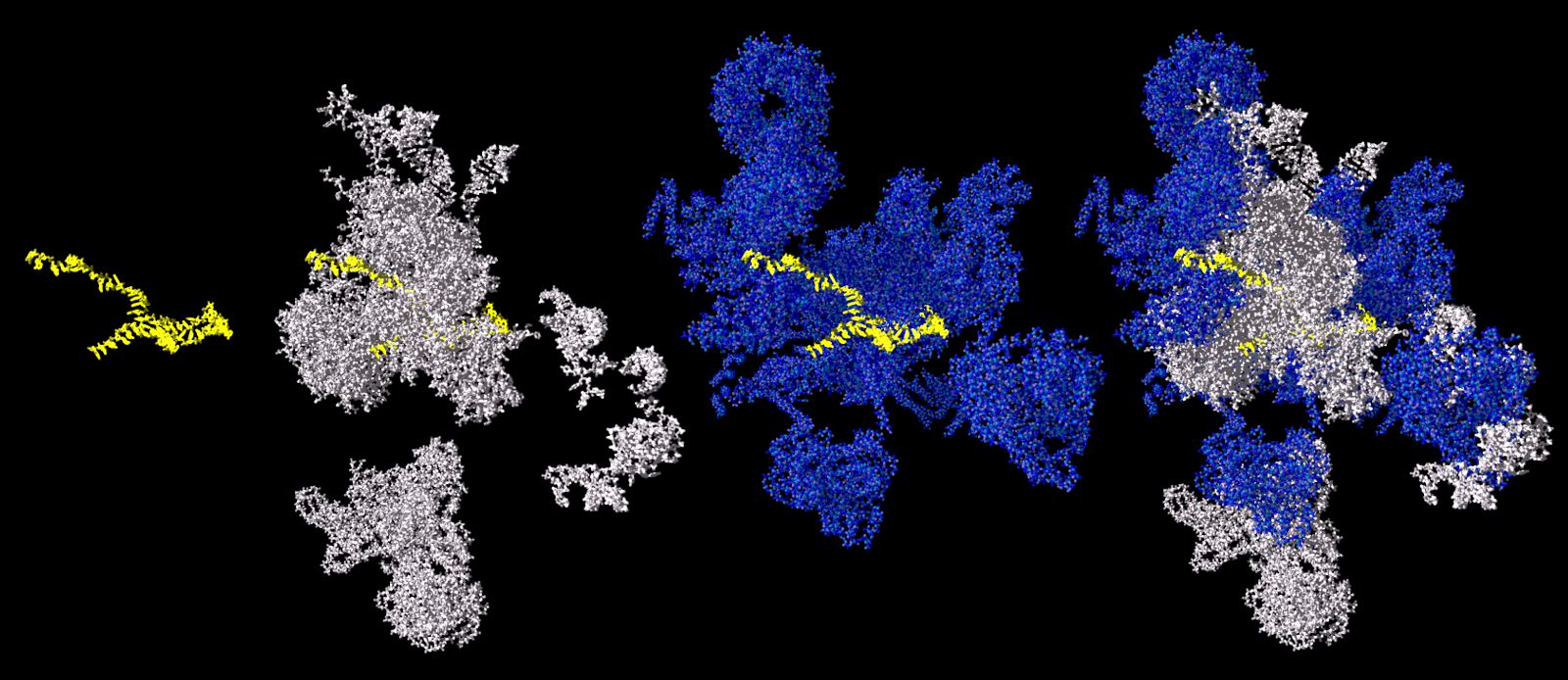
The figure shows a spliceosome after its first catalytic step (as described by Galej et al.) and partially decomposed in its various components. The figure has been adapted from an entry (5LJ5) in the Protein Data Bank. The RNA to be spliced is given in yellow colour, the RNA-part of the spliceosome in white colour and the protein-part of the spliceosome in blue colour.
Source: Galej et al., Nature 537, 197 – 203 // Rauhut et al., Science 353, 1399 – 1405