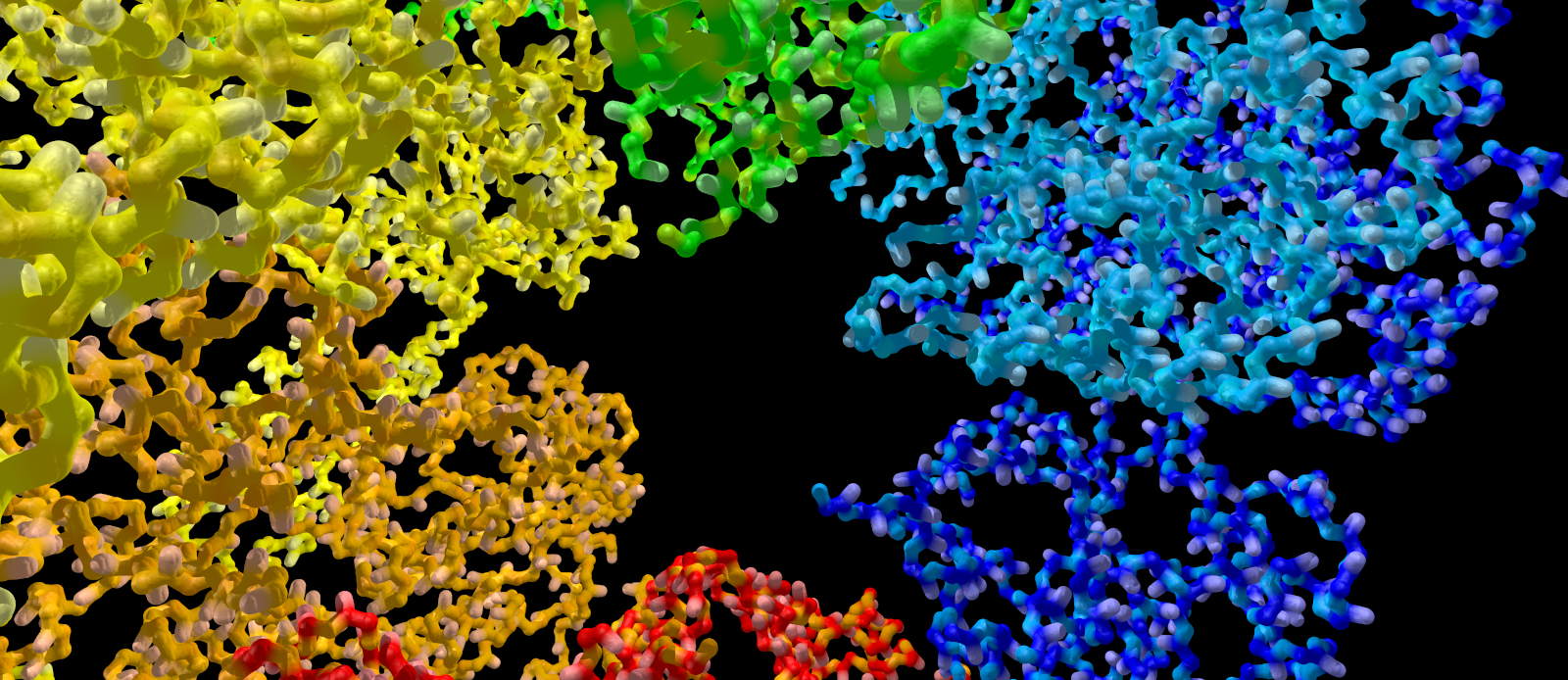
Protein disaggregation by a spiral enzyme
Under cellular stress protein disaggregation becomes an important issue for living cells. Enzymes involved in this job therefore often have been found as heat shock proteins. The heat shock protein 104 from yeast is a good example for such enzymes. As many other, so called AAA+ disaggregases, it untangles protein aggregates while consuming ATP. When studying its structures scientists from the US observed a surprising novel architecture for this class of enzymes.
For elucidating the enzyme’s structure yeast Hsp 104 was bound to non-hydrolyzable ATP and studied using Cryo-EM. While many other AAA+ disaggregases are known as ring-like structures, Hsp 104 hexamers were observed to form spirals. In the figure accompanying this short summary, each monomer of this spiral has been given a different colour. Each of the monomers has two ATP-binding sites which are forming a double spiral structure in the central space of the assembled enzyme. It seems likely that this double spiral arrangement describes the path of the substrate polypeptide across the disaggregase.
Source: Yokom et al., Nature Structural & Molecular Biology doi:10.1038/nsmb.3277