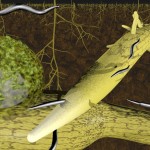
DNA-molecules can become very long (up to several centimetres) while having a very small diameter (about 2.5 nanometres in the case of the double helix). As a consequence, they can be regarded as ordered structures connecting the macroscopic range and the nano-range.
Packaging of such molecules (within cells or cell nuclei) is a very interesting topic, first because packaging determines, how fast the information stored in the DNA can be used by the cell, and second, because cellular compartments appear to be tiny compared to DNA-molecules. The following two examples compare the size of DNA-molecules to the size of respective cellular compartments.
The genome of the model bacterium Escherichia coli has a size of 4.600.000 base pairs, which translates into a length of the circular DNA-molecules of 1.518.000 nanometres or 1.5 millimetres. A typical Escherichia coli containing one such molecule (or two molecules prior to cell division) is a few micrometers long, i.e., almost 1/1000 the size of these molecules.
The human genome has a size of 3.200.000.000 base pairs, which translates into a total length of 1.056.000.000 nanometres or 1.06 meters. The genome is partitioned into 23 portions (the chromosomes) with a size of about 4.6 centimetres each. Each nucleus of our body cells has a diameter of a few micrometers and contains either 2 or 4 such copies (depending on its stage in the cell cycle), it means 2 or 4 meters of DNA-double helices. This means the size of the nucleus is 1/millionth the size of the molecules; how do they fit inside?
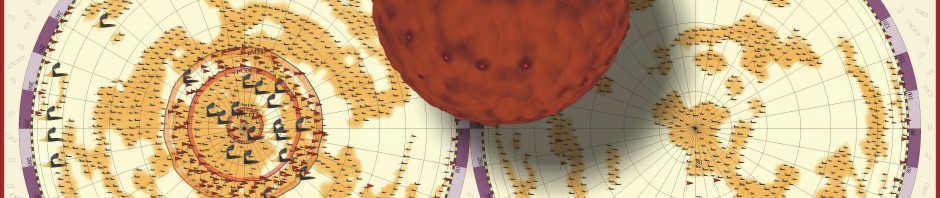
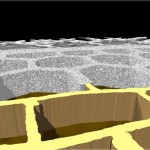
Part of the answer is given by the small diameter of DNA-molecules. Due to this diameter even very long molecules occupy only a small volume. This is easily shown by cutting the DNA-molecules into small fragments of equal size and aligning them one by one closely together, as demonstrated below for the genome from Escherichia coli. (Each cylinder represents a DNA-double helix).

E. coli: Genomic square
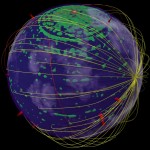
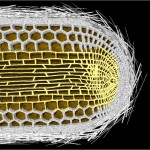
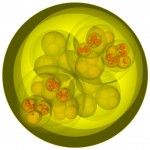
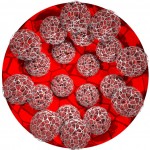
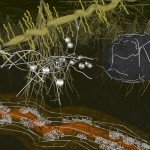
In the case of Escherichia coli such a genomic square is composed of 779 fragments of DNA with an edge length of 1948 micrometers, in the case of our own DNA the genomic square is composed of 35777 fragments with an edge length of 89442 micrometers (this won’t work on my computer). As demonstrated in the following figures, the genomic square still overlaps Escherichia coli cells, but packaging of the material does not seem to be a major problem.

E.coli and its genome
E. coli and its genome (front view)
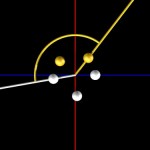
So packaging seems doable, but how is it really done. Here, I will only present one recurring motif of DNA-packaging, which is the use of fractal loops (or helices, or coils). These are exemplified below with molecules 8.500 base pairs long.
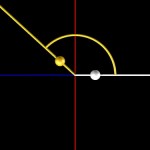
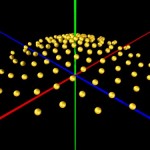
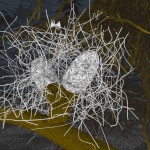
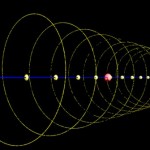
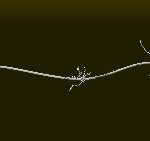
The helical conformation of the DNA double helix already shortens the molecules considerably (with the calculations above referring to such double helical molecules, however).

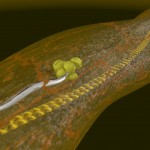
DNA double helix
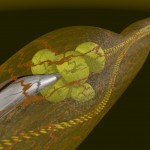
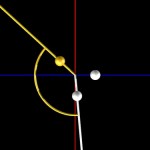
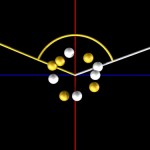
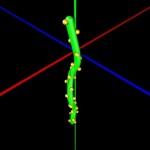
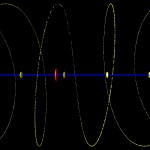
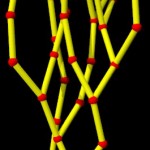
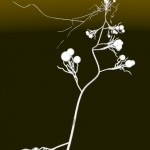
Double helices can be arranged into loops.

looped DNA
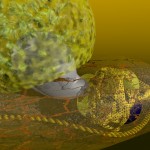
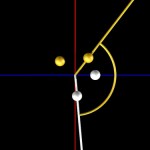
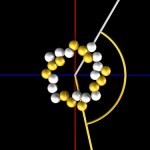
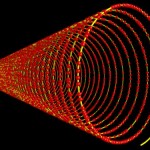
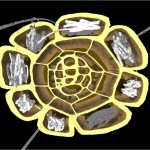
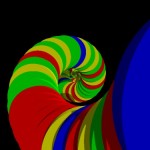
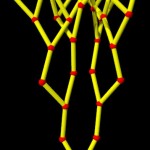
And these, again, into super-loops, which, of course, is not the last level of looping.
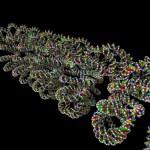
super-looped DNA
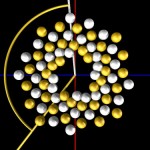
Nota bene: This is only a concept, the reality is much more complex.
Anyway, these pictures should give an idea, how very long molecules fit into very small spaces.
There is only one geometric mystery remaining (at least to me), connected to the replication of DNA. Replication occurs within the nucleus, so the bulk of DNA has to stay packaged while small segments are unpacked to become replicated. In the end the newly formed molecules are condensed even further (forming chromosomes), to allow the spatial separation of the two copies. How is it possible that the newly formed molecules are not hopelessly knotted? Does anybody have an idea?
Further reading:
Histones: DNA packaging and much more
Scientists unwrap DNA packaging to gain insight into cells
DNA Chromosome packaging animation
A cellular secret to long life